Biochemistry
|
9 december 2018 12:00:06 |
| Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain (Structure) |
|
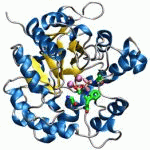
Tweet Lee et al. assess the druggability of the ionotropic glutamate receptor subfamilies, using molecular dynamics simulations in the presence of drug-like molecules and X-ray crystallography. The study presents a ligand-binding site in the GluA3 N-terminal domain and supports the role of conformational plasticity in modulating ligand binding. |
| 106 viewsCategory: Biochemistry |
 Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155 (Structure) Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155 (Structure)Computational Prediction of Amino Acids Governing Protein-Membrane Interaction for the PIP3 Cell Signaling System (Structure) 
|
| blog comments powered by Disqus |
MyJournals.org
The latest issues of all your favorite science journals on one page
The latest issues of all your favorite science journals on one page